Convergence tests
It is good idea to check the convergence of total energy with respect various parameters of our calculation e.g., kinetic energy cutoff, k-grid density, lattice constant etc.
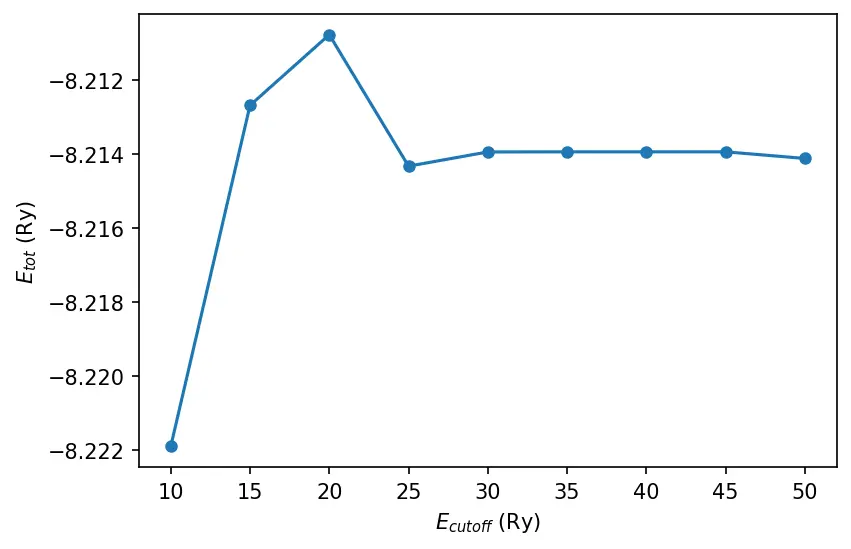
Cutoff energy
Here is shell script example to programmatically create input files and run calculations while varying certain parameter:
src/convergence/ecut.sh
#!/bin/sh
NAME="ecut"
for CUTOFF in `seq 10 5 50`
do
cat > ${NAME}_${CUTOFF}.in << EOF
System.CurrrentDirectory ./
DATA.PATH /workspaces/openmx3.9/DFT_DATA19
System.Name Si_ecut_${CUTOFF}
Species.Number 1
<Definition.of.Atomic.Species
Si Si7.0-s2p2d1 Si_PBE19
Definition.of.Atomic.Species>
Atoms.Number 2
Atoms.SpeciesAndCoordinates.Unit Ang # Ang|AU
<Atoms.SpeciesAndCoordinates # Unit=Ang.
1 Si 0.000000000000 0.000000000000 0.000000000000 2.0 2.0
2 Si 1.357500000000 1.357500000000 1.357500000000 2.0 2.0
Atoms.SpeciesAndCoordinates>
Atoms.UnitVectors.Unit Ang # Ang|AU
<Atoms.UnitVectors # unit=Ang.
0.000000000000 2.715000000000 2.715000000000
2.715000000000 0.000000000000 2.715000000000
2.715000000000 2.715000000000 0.000000000000
Atoms.UnitVectors>
scf.XcType GGA-PBE # LDA|LSDA-CA|LSDA-PW
scf.SpinPolarization Off # On|Off|NC
scf.SpinOrbit.Coupling Off # On|Off, default=off
scf.ElectronicTemperature 300.0 # default=300 (K)
scf.energycutoff ${CUTOFF} # default=150 (Ry)
scf.maxIter 200 # default=40
scf.EigenvalueSolver band # Recursion|Cluster|Band
scf.lapack.dste dstevx # dstegr|dstedc|dstevx, default=dstegr
scf.Kgrid 5 5 5 # means 4x4x4
scf.Mixing.Type rmm-diisk # Simple|Rmm-Diis|Gr-Pulay
scf.Init.Mixing.Weight 0.010 # default=0.30
scf.Min.Mixing.Weight 0.001 # default=0.001
scf.Max.Mixing.Weight 0.200 # default=0.40
scf.Mixing.History 15 # default=5
scf.Mixing.StartPulay 5 # default=6
scf.criterion 1.0e-9 # default=1.0e-6 (Hartree)
EOF
echo "Calculating ${NAME}_${CUTOFF}"
mpirun -np 4 /workspaces/openmx3.9/work/openmx ${NAME}_${CUTOFF}.in > ${NAME}_${CUTOFF}.out
grep -E "Utot\s+=" ${NAME}_${CUTOFF}.out
done
The script will print out total energy for each cutoff energy used. We can plot the result:
import numpy as np
import matplotlib.pyplot as plt
%matplotlib inline
plt.rcParams["figure.dpi"]=150
plt.rcParams["figure.facecolor"]="white"
ecut, etot = np.loadtxt("../src/convergence/ecut-vs-etot.dat", unpack=True)
plt.plot(ecut, etot, "o-", markersize=5)
plt.xlabel("$E_{cutoff}$ (Ry)")
plt.ylabel("$E_{tot}$ (Ry)")
plt.show()
note
The cutoff energy in OpenMX is not for the basis set as in plane wave methods, but for the numerical integrations. Therefore the total energy does not have to converge from the upper energy region with respect to the cutoff energy like that of plane wave basis set.
k-grid density
src/convergence/kgrid.sh
#!/bin/sh
NAME="Kgrid"
for GRID in `seq 2 10`
do
cat > ${NAME}_${GRID}.in << EOF
System.CurrrentDirectory ./
DATA.PATH /workspaces/openmx3.9/DFT_DATA19
System.Name Si_${GRID}
Species.Number 1
<Definition.of.Atomic.Species
Si Si7.0-s2p2d1 Si_PBE19
Definition.of.Atomic.Species>
Atoms.Number 2
Atoms.SpeciesAndCoordinates.Unit Ang # Ang|AU
<Atoms.SpeciesAndCoordinates # Unit=Ang.
1 Si 0.000000000000 0.000000000000 0.000000000000 2.0 2.0
2 Si 1.357500000000 1.357500000000 1.357500000000 2.0 2.0
Atoms.SpeciesAndCoordinates>
Atoms.UnitVectors.Unit Ang # Ang|AU
<Atoms.UnitVectors # unit=Ang.
0.000000000000 2.715000000000 2.715000000000
2.715000000000 0.000000000000 2.715000000000
2.715000000000 2.715000000000 0.000000000000
Atoms.UnitVectors>
scf.XcType GGA-PBE # LDA|LSDA-CA|LSDA-PW
scf.SpinPolarization Off # On|Off|NC
scf.SpinOrbit.Coupling Off # On|Off, default=off
scf.ElectronicTemperature 300.0 # default=300 (K)
scf.energycutoff 50 # default=150 (Ry)
scf.maxIter 200 # default=40
scf.EigenvalueSolver band # Recursion|Cluster|Band
scf.lapack.dste dstevx # dstegr|dstedc|dstevx, default=dstegr
scf.Kgrid ${GRID} ${GRID} ${GRID} # means 4x4x4
scf.Mixing.Type rmm-diisk # Simple|Rmm-Diis|Gr-Pulay
scf.Init.Mixing.Weight 0.010 # default=0.30
scf.Min.Mixing.Weight 0.001 # default=0.001
scf.Max.Mixing.Weight 0.200 # default=0.40
scf.Mixing.History 15 # default=5
scf.Mixing.StartPulay 5 # default=6
scf.criterion 1.0e-9 # default=1.0e-6 (Hartree)
EOF
echo "Calculating ${NAME}_${GRID}"
mpirun -np 4 /workspaces/openmx3.9/work/openmx ${NAME}_${GRID}.in > ${NAME}_${GRID}.out
grep -E "Utot\s+=" ${NAME}_${GRID}.out
done
k, etot = np.loadtxt("../src/convergence/kgrid-vs-etot.dat", unpack=True)
plt.plot(k, etot, "o-", markersize=5)
plt.xlabel("k_grid")
plt.ylabel("$E_{tot}$ (Ry)")
plt.show()
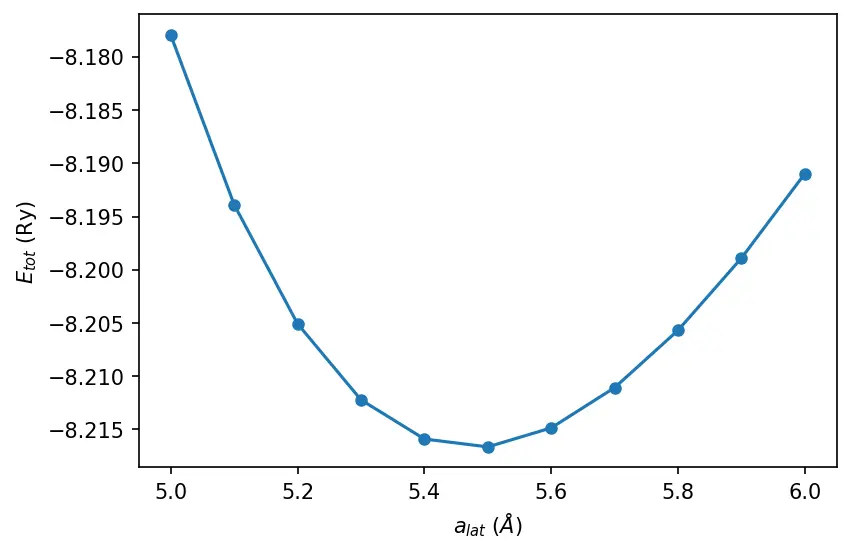
Lattice constant
src/convergence/alat.sh
#!/bin/sh
NAME="alat"
for ALAT in `seq 5 0.1 6`
do
ALAT2=$(echo "scale=7;${ALAT}/2" | bc)
ALAT4=$(echo "scale=7;${ALAT}/4" | bc)
cat > ${NAME}_${ALAT}.in << EOF
System.CurrrentDirectory ./
DATA.PATH /workspaces/openmx3.9/DFT_DATA19
System.Name Si_${ALAT}
Species.Number 1
<Definition.of.Atomic.Species
Si Si7.0-s2p2d1 Si_PBE19
Definition.of.Atomic.Species>
Atoms.Number 2
Atoms.SpeciesAndCoordinates.Unit Ang # Ang|AU
<Atoms.SpeciesAndCoordinates # Unit=Ang.
1 Si 0.000000000000 0.000000000000 0.000000000000 2.0 2.0
2 Si ${ALAT4} ${ALAT4} ${ALAT4} 2.0 2.0
Atoms.SpeciesAndCoordinates>
Atoms.UnitVectors.Unit Ang # Ang|AU
<Atoms.UnitVectors # unit=Ang.
0.000000000000 ${ALAT2} ${ALAT2}
${ALAT2} 0.000000000000 ${ALAT2}
${ALAT2} ${ALAT2} 0.000000000000
Atoms.UnitVectors>
scf.XcType GGA-PBE # LDA|LSDA-CA|LSDA-PW
scf.SpinPolarization Off # On|Off|NC
scf.SpinOrbit.Coupling Off # On|Off, default=off
scf.ElectronicTemperature 300.0 # default=300 (K)
scf.energycutoff 50 # default=150 (Ry)
scf.maxIter 200 # default=40
scf.EigenvalueSolver band # Recursion|Cluster|Band
scf.lapack.dste dstevx # dstegr|dstedc|dstevx, default=dstegr
scf.Kgrid 8 8 8 # means 4x4x4
scf.Mixing.Type rmm-diisk # Simple|Rmm-Diis|Gr-Pulay
scf.Init.Mixing.Weight 0.010 # default=0.30
scf.Min.Mixing.Weight 0.001 # default=0.001
scf.Max.Mixing.Weight 0.200 # default=0.40
scf.Mixing.History 15 # default=5
scf.Mixing.StartPulay 5 # default=6
scf.criterion 1.0e-9 # default=1.0e-6 (Hartree)
EOF
echo "Calculating ${NAME}_${ALAT}"
mpirun -np 4 /workspaces/openmx3.9/work/openmx ${NAME}_${ALAT}.in > ${NAME}_${ALAT}.out
grep -E "Utot\s+=" ${NAME}_${ALAT}.out
done
alat, etot = np.loadtxt("../src/convergence/alat-vs-etot.dat", unpack=True)
plt.plot(alat, etot, "o-", markersize=5)
plt.xlabel("$a_{lat}$ ($\\AA$)")
plt.ylabel("$E_{tot}$ (Ry)")
plt.show()